Chapter 6: Single group studies using SAS
Figure 6.1 Output for calculating a prevalence and 95% confidence interval
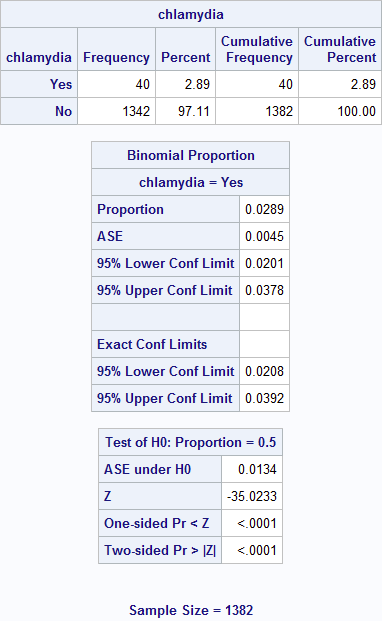
Figure 6.1
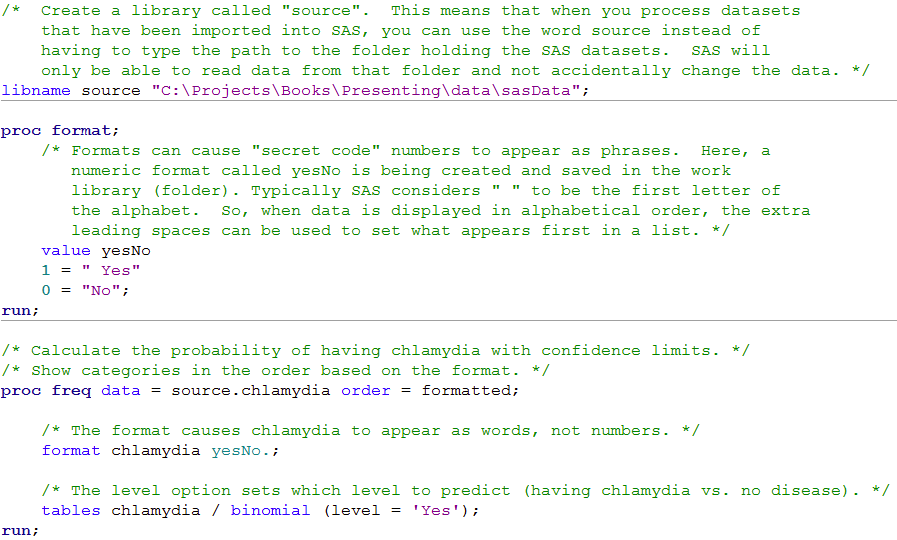
/* Create a library called "source". This means that when you process datasets
that have been imported into SAS, you can use the word source instead of
having to type the path to the folder holding the SAS datasets. SAS will
only be able to read data from that folder and not accidentally change the data. */
libname source "C:\Projects\Books\Presenting\data\sasData";
proc format;
/* Formats can cause "secret code" numbers to appear as phrases. Here, a
numeric format called yesNo is being created and saved in the work
library (folder). Typically SAS considers " " to be the first letter of
the alphabet. So, when data is displayed in alphabetical order, the extra
leading spaces can be used to set what appears first in a list. */
value yesNo
1 = " Yes"
0 = "No";
run;
/* Calculate the probability of having chlamydia with confidence limits. */
/* Show categories in the order based on the format. */
proc freq data = source.chlamydia order = formatted;
/* The format causes chlamydia to appear as words, not numbers. */
format chlamydia yesNo.;
/* The level option sets which level to predict (having chlamydia vs. no disease). */
tables chlamydia / binomial (level = 'Yes');
run;
Figure 6.2 Calculating sensitivity and specificity
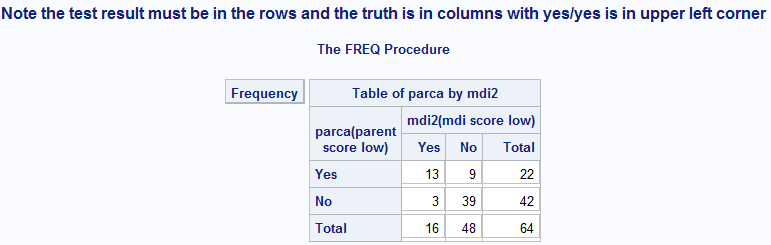
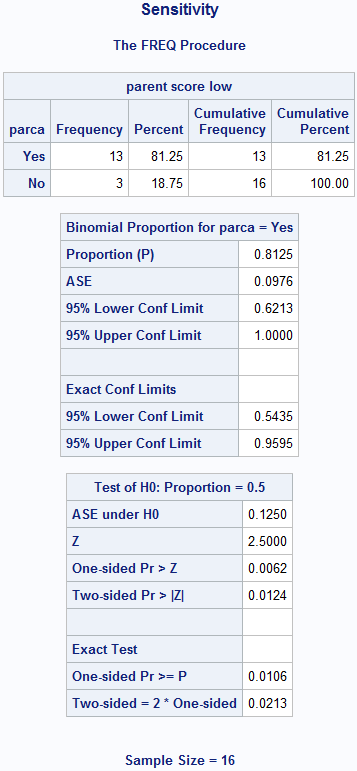
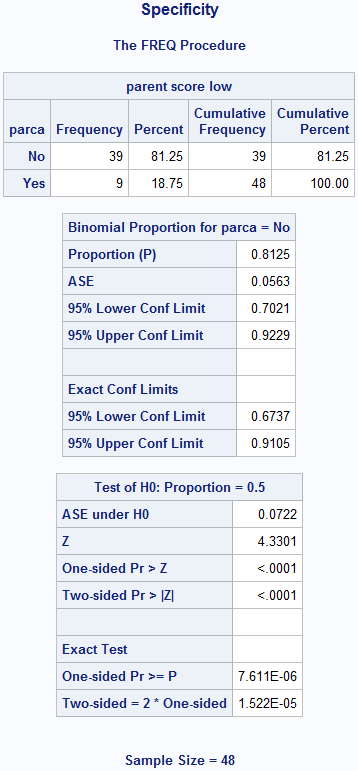
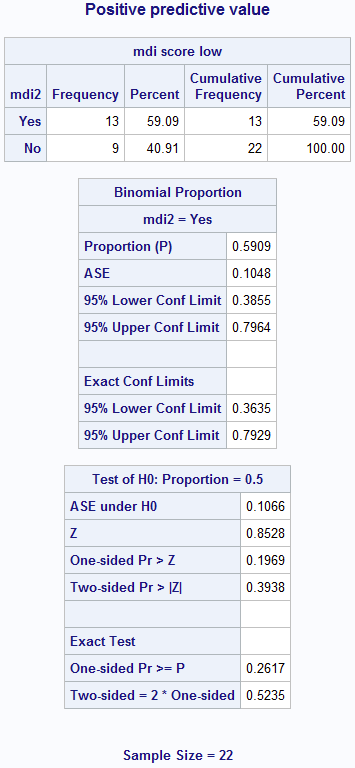
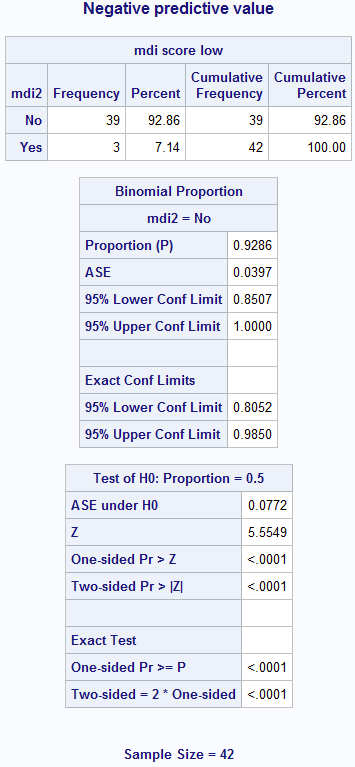
Figure 6.2 Code
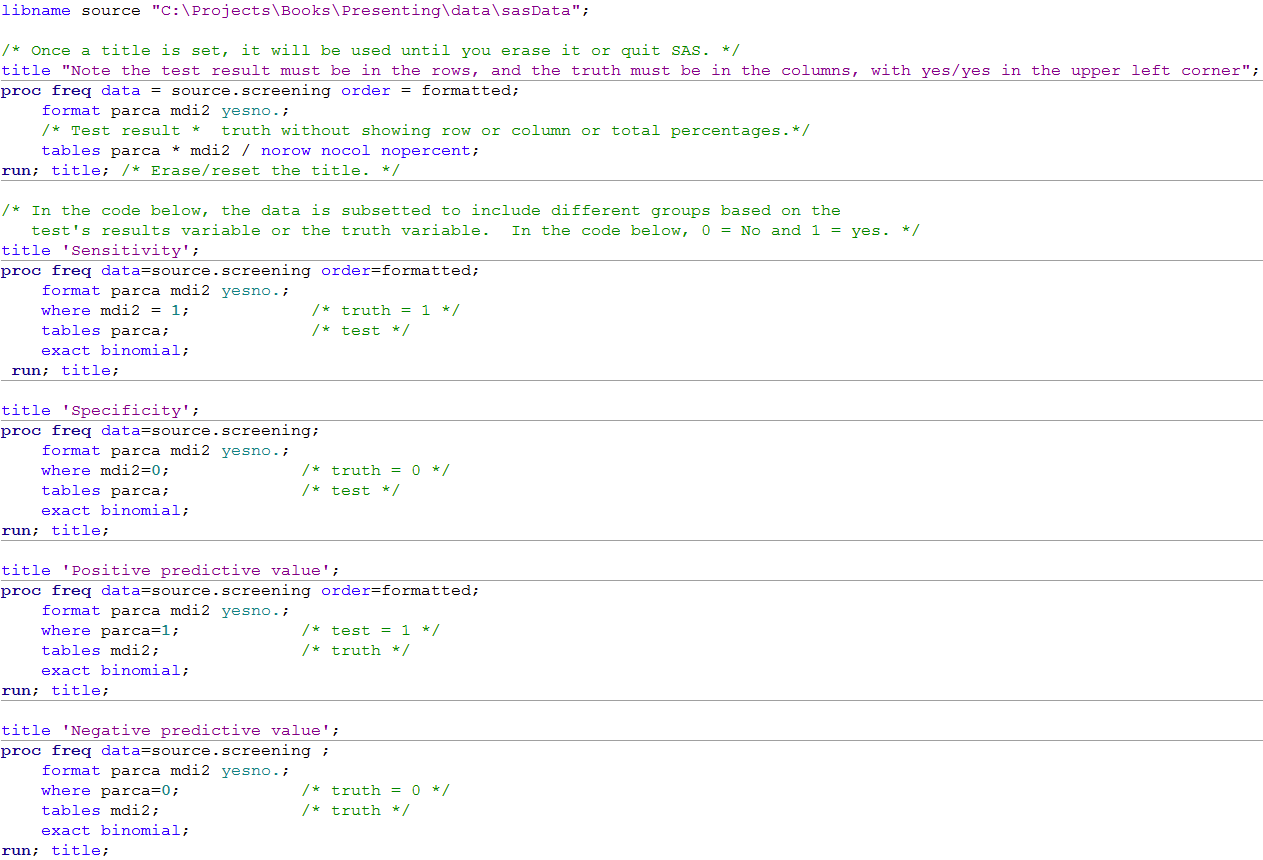 Click here to show code as text
Click here to show code as text
libname source "C:\Projects\Books\Presenting\data\sasData";
/* Once a title is set, it will be used until you erase it or quit SAS. */
title "Note the test result must be in the rows, and the truth must be in the columns, with yes/yes in the upper left corner";
proc freq data = source.screening order = formatted;
format parca mdi2 yesno.;
/* Test result * truth without showing row or column or total percentages.*/
tables parca * mdi2 / norow nocol nopercent;
run; title; /* Erase/reset the title. */
/* In the code below, the data is subsetted to include different groups based on the
test's results variable or the truth variable. In the code below, 0 = No and 1 = yes. */
title 'Sensitivity';
proc freq data=source.screening order=formatted;
format parca mdi2 yesno.;
where mdi2 = 1; /* truth = 1 */
tables parca; /* test */
exact binomial;
run; title;
title 'Specificity';
proc freq data=source.screening;
format parca mdi2 yesno.;
where mdi2=0; /* truth = 0 */
tables parca; /* test */
exact binomial;
run; title;
title 'Positive predictive value';
proc freq data=source.screening order=formatted;
format parca mdi2 yesno.;
where parca=1; /* test = 1 */
tables mdi2; /* truth */
exact binomial;
run; title;
title 'Negative predictive value';
proc freq data=source.screening ;
format parca mdi2 yesno.;
where parca=0; /* truth = 0 */
tables mdi2; /* truth */
exact binomial;
run; title;
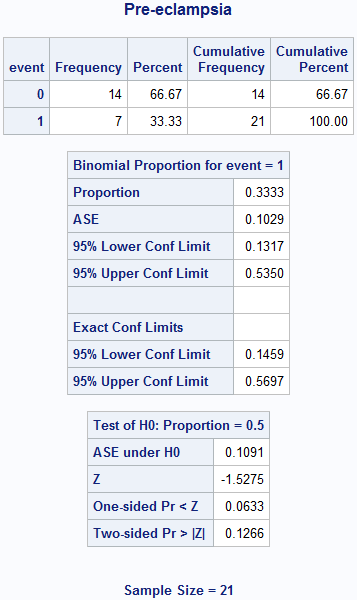
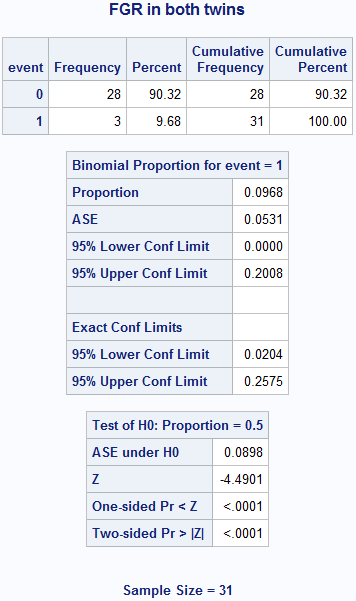
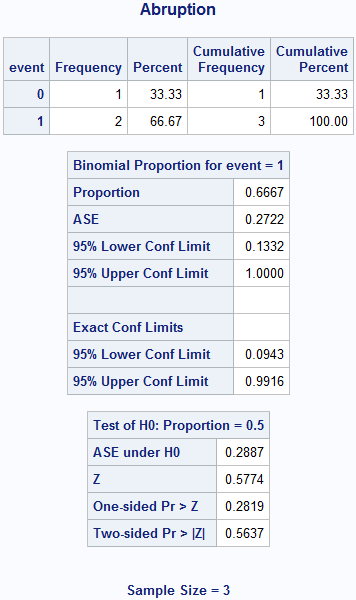
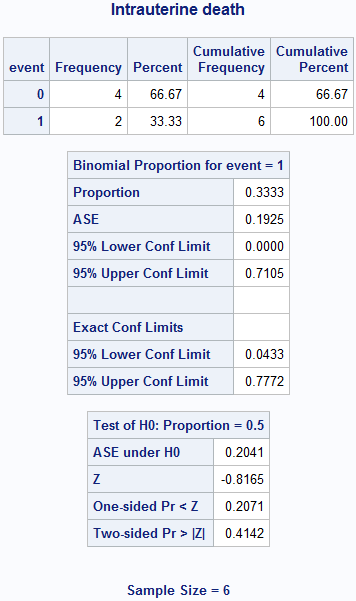
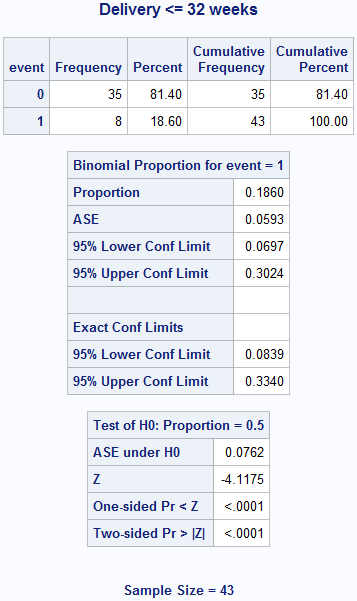
Box 6.3 Screening study with a rare condition
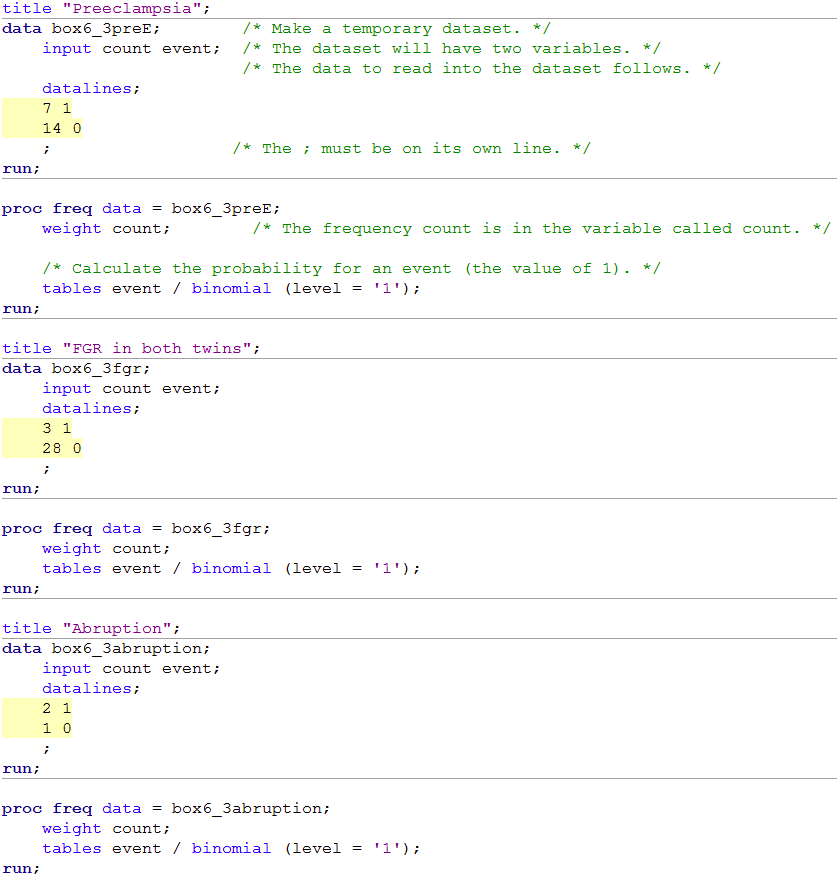
Box 6.3 Code
 Click here to show code as text
Click here to show code as text
title "Preeclampsia";
data box6_3preE; /* Make a temporary dataset. */
input count event; /* The dataset will have two variables. */
/* The data to read into the dataset follows. */
datalines;
7 1
14 0
; /* The ; must be on its own line. */
run;
proc freq data = box6_3preE;
weight count; /* The frequency count is in the variable called count. */
/* Calculate the probability for an event (the value of 1). */
tables event / binomial (level = '1');
run;
title "FGR in both twins";
data box6_3fgr;
input count event;
datalines;
3 1
28 0
;
run;
proc freq data = box6_3fgr;
weight count;
tables event / binomial (level = '1');
run;
title "Abruption";
data box6_3abruption;
input count event;
datalines;
2 1
1 0
;
run;
proc freq data = box6_3abruption;
weight count;
tables event / binomial (level = '1');
run;
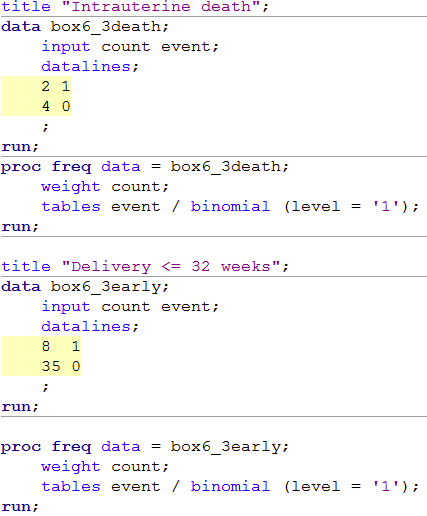
title "Intrauterine death";
data box6_3death;
input count event;
datalines;
2 1
4 0
;
run;
proc freq data = box6_3death;
weight count;
tables event / binomial (level = '1');
run;
title "Delivery <= 32 weeks";
data box6_3early;
input count event;
datalines;
8 1
35 0
;
run;
proc freq data = box6_3early;
weight count;
tables event / binomial (level = '1');
run;